lorentzian_diff¶
- Stoner.analysis.fitting.models.generic.lorentzian_diff(x, A, sigma, mu)[source]¶
Implement a differential form of a Lorentzian peak.
- Parameters:
x (array) – x data
A (float) – Peak amplitude
sigma (float) – peak wideth
mu (float) – peak location in x
- Returns
- :math:`frac{A sigma left(2 mu - 2 xright)}{pi left(sigma^{2} +
left(- mu + xright)^{2}right)^{2}}`
Example
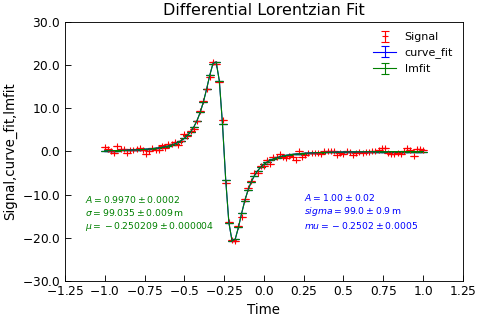
"""Test langevin fitting.""" from copy import copy from numpy import linspace, ones_like from numpy.random import normal from Stoner import Data from Stoner.analysis.fitting.models.generic import ( Lorentzian_diff, lorentzian_diff, ) x = linspace(-1.0, 1.0, 101) params = [1, 0.1, -0.25] y = lorentzian_diff(x, *params) + normal(size=len(x), scale=0.5) dy = ones_like(x) * 5e-3 d = Data(x, y, dy, setas="xye", column_headers=["Time", "Signal", "dM"]) d.curve_fit(lorentzian_diff, p0=copy(params), result=True, header="curve_fit") d.setas = "xye" d.lmfit(Lorentzian_diff, result=True, header="lmfit", prefix="lmfit") d.setas = "xyeyy" d.plot(fmt=["r+", "b-", "g-"]) d.annotate_fit( lorentzian_diff, x=0.6, y=0.2, fontdict={"size": "x-small", "color": "blue"}, mode="eng", ) d.annotate_fit( Lorentzian_diff, x=0.05, y=0.2, fontdict={"size": "x-small", "color": "green"}, prefix="lmfit", mode="eng", ) d.title = "Differential Lorentzian Fit"