Data.differential_evolution¶
- Data.differential_evolution(model, xcol=None, ycol=None, p0=None, sigma=None, **kargs)¶
Fit model to the data using a differential evolution algorithm.
- Parameters:
model (lmfit.Model) – An instance of an lmfit.Model that represents the model to be fitted to the data
xcol (index or None) – Columns to be used for the x data for the fitting. If not givem defaults to the
Stoner.Core.DataFile.setasx columnycol (index or None) – Columns to be used for the y data for the fitting. If not givem defaults to the
Stoner.Core.DataFile.setasy column
- Keyword Arguments:
p0 (list, tuple, array or callable) – A vector of initial parameter values to try. See the notes in
Stoner.Data.curve_fit()for more details.sigma (index) – The index of the column with the y-error bars
bounds (callable) – A callable object that evaluates true if a row is to be included. Should be of the form f(x,y)
result (bool) – Determines whether the fitted data should be added into the DataFile object. If result is True then the last column will be used. If result is a string or an integer then it is used as a column index. Default to None for not adding fitted data
replace (bool) – Inidcatesa whether the fitted data replaces existing data or is inserted as a new column (default False)
header (string or None) – If this is a string then it is used as the name of the fitted data. (default None)
scale_covar (bool) – whether to automatically scale covariance matrix (leastsq only)
output (str, default "fit") – Specify what to return.
- Returns:
( various ) –
- The return value is determined by the output parameter. Options are
- ”fit” just the
lmfit.model.ModelFitinstance that contains all relevant information about the fit.
- ”fit” just the
- ”row” just a one dimensional numpy array of the fit parameters interleaved with their
uncertainties
”full” a tuple of the fit instance and the row.
- ”data” a copy of the
Stoner.Core.DataFileobject with the fit recorded in the emtadata and optionally as a column of data.
- ”data” a copy of the
This function is essentially a wrapper around the
scipy.optimize.differential_evolution()function that presents the same interface as the other Stoner package curve fitting functions. The parent function, however, does not provide the variance-covariance matrix to estimate the fitting errors. To work around this, this function does the initial fit with the differential evolution, but then uses that to give a starting vector to a call toscipy.optimize.curve_fit()to calculate the covariance matrix.See also
Example
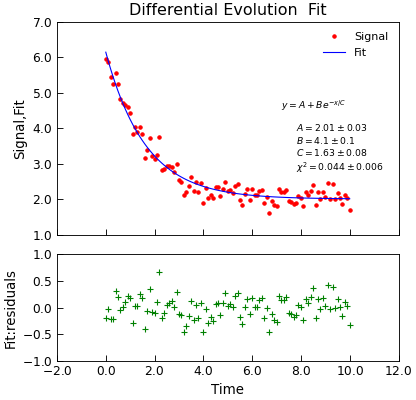
"""Simple use of lmfit to fit data.""" from numpy import linspace, exp, random from Stoner import Data random.seed(12345) # Ensure consistent random numbers! # Make some data x = linspace(0, 10.0, 101) y = 2 + 4 * exp(-x / 1.7) + random.normal(scale=0.2, size=101) d = Data(x, y, column_headers=["Time", "Signal"], setas="xy") func = lambda x, A, B, C: A + B * exp(-x / C) # Do the fitting and plot the result fit = d.differential_evolution( func, result=True, header="Fit", A=1, B=1, C=1, prefix="Model", residuals=True, ) # Reset labels d.labels = [] # Make nice two panel plot layout ax = d.subplot2grid((3, 1), (2, 0)) d.setas = "x..y" d.plot(fmt="g+") d.title = "" ax = d.subplot2grid((3, 1), (0, 0), rowspan=2) d.setas = "xyy" d.plot(fmt=["r.", "b-"]) d.xticklabels = [[]] d.xlabel = "" # Annotate plot with fitting parameters d.annotate_fit(func, prefix="Model", x=0.7, y=0.3, fontdict={"size": "x-small"}) text = r"$y=A+Be^{-x/C}$" + "\n\n" d.text(7.2, 3.9, text, fontdict={"size": "x-small"}) d.title = "Differential Evolution Fit"