HGXFile¶
- class Stoner.HDF5.HGXFile(*args)[source]¶
Bases:
DataFileA subclass of DataFile for reading GenX HDF Files.
These files typically have an extension .hgx. This class has been based on a limited sample of hgx files and so may not be sufficiently general to handle all cases.
Attributes Summary
Get the current data transposed.
Return the basename of the current filename.
Get a deep copy of the current DataFile.
Pass through to the setas attribute.
Property Accessors for the main numerical data.
Return the data as a dictionary of single columns with column headers for the keys.
Alias for self.data.axes.
Return the np dtype attribute of the data.
Return DataFile filename, or make one up.
Return DataFile filename, or make one up, returning as a pathlib.Path.
Make a pretty header string that looks like the tabular representation.
Return the mask of the data array.
Read the metadata dictionary.
Return the data as a np structured data array.
Get the list of column assignments.
Pass through the numpy shape attribute of the data.
Methods Summary
__call__(*args, **kargs)Clone the DataFile, but allowing additional arguments to modify the new clone.
add_column(column_data[, header, index, ...])Append a column of data or inserts a column to a datafile instance.
append(value)S.append(value) -- append value to the end of the sequence
asarray()Provide a consistent way to get at the underlying array data.
clear()closest(value[, xcol])Return the row in a data file which has an x-column value closest to the given value.
column(col)Extract one or more columns of data from the datafile.
columns([not_masked, reset])Iterate over the columns of data int he datafile.
count([value, axis, col])Count the number of un-masked elements in the
DataFile.del_column([col, duplicates])Delete a column from the current
DataFileobject.del_nan([col, clone])Remove rows that have nan in them.
del_rows([col, val, invert])Search in the numerica data for the lines that match and deletes the corresponding rows.
dir([pattern])Return a list of keys in the metadata, filtering with a regular expression if necessary.
extend(values)S.extend(iterable) -- extend sequence by appending elements from the iterable
filter([func, cols, reset])Set the mask on rows of data by evaluating a function for each row.
find_col(col[, force_list])Indexes the column headers in order to locate a column of data.shape.
find_duplicates([xcol, delta])Find rows with duplicated values of the search column(s).
get(k[,d])get_filename(mode)Force the user to choose a new filename using a system dialog box.
index(value, [start, [stop]])Raises ValueError if the value is not present.
insert(index, obj)Implement the insert method.
insert_rows(row, new_data)Insert new_data into the data array at position row.
items()Make sure we implement an items that doesn't just iterate over self.
keys()Return the keys of the metadata dictionary.
load(*args, **kargs)Create a new
DataFilefrom a file on disc guessing a better subclass if necessary.main_data(data_grp)Work through the main data group and build something that looks like a numpy 2D array.
pop(k[,d])If key is not found, d is returned if given, otherwise KeyError is raised.
popitem()as a 2-tuple; but raise KeyError if D is empty.
remove(value)S.remove(value) -- remove first occurrence of value.
remove_duplicates([xcol, delta, strategy, ...])Find and remove rows with duplicated values of the search column(s).
rename(old_col, new_col)Rename columns without changing the underlying data.
reorder_columns(cols[, headers_too, setas_too])Construct a new data array from the original data by assembling the columns in the order given.
reverse()S.reverse() -- reverse IN PLACE
rolling_window([window, wrap, exclude_centre])Iterate with a rolling window section of the data.
rows([not_masked, reset])Iterate over rows of data.
save([filename])Save a string representation of the current DataFile object into the file 'filename'.
scan_group(grp, pth)Recursively list HDF5 Groups.
search([xcol, value, columns, accuracy])Search the numerica data part of the file for lines that match and returns the corresponding rows.
section(**kargs)Assuming data has x,y or x,y,z coordinates, return data from a section of the parameter space.
select(*args, **kargs)Produce a copy of the DataFile with only data rows that match a criteria.
setdefault(k[,d])sort(*order, **kargs)Sort the data by column name.
split(*args[, final])Recursively splits the current DataFile into a
Stoner.Folders.DataFolder.swap_column(*swp, **kargs)Swap pairs of columns in the data.
Create a pandas DataFrame from a
Stoner.Dataobject.unique(col[, return_index, return_inverse])Return the unique values from the specified column - pass through for numpy.unique.
update([E, ]**F)If E present and has a .keys() method, does: for k in E: D[k] = E[k] If E present and lacks .keys() method, does: for (k, v) in E: D[k] = v In either case, this is followed by: for k, v in F.items(): D[k] = v
values()Return the values of the metadata dictionary.
Attributes Documentation
- T¶
Get the current data transposed.
- basename¶
Return the basename of the current filename.
- clone¶
Get a deep copy of the current DataFile.
- column_headers¶
Pass through to the setas attribute.
- data¶
Property Accessors for the main numerical data.
- dict_records¶
Return the data as a dictionary of single columns with column headers for the keys.
- dims¶
Alias for self.data.axes.
- dtype¶
Return the np dtype attribute of the data.
- filename¶
Return DataFile filename, or make one up.
- filepath¶
Return DataFile filename, or make one up, returning as a pathlib.Path.
- header¶
Make a pretty header string that looks like the tabular representation.
- mask¶
Return the mask of the data array.
- metadata¶
Read the metadata dictionary.
- mime_type = ['application/x-hdf', 'application/x-hdf5']¶
- pattern = ['*.hgx']¶
- patterns = ['*.txt', '*.tdi', '*.hdf', '*.hf5', '*.zip']¶
- priority = 16¶
- records¶
Return the data as a np structured data array.
If columns names are duplicated then they are made unique.
- setas¶
Get the list of column assignments.
- shape¶
Pass through the numpy shape attribute of the data.
Methods Documentation
- __call__(*args, **kargs)¶
Clone the DataFile, but allowing additional arguments to modify the new clone.
- Parameters:
*args (tuple) – Positional arguments to pass through to the new clone.
**kargs (dict) – Keyword arguments to pass through to the new clone.
- Raises:
TypeError – If a keyword argument doesn’t match an attribute.
- Returns:
new_d (DataFile) – Modified clone of the current object.
- add_column(column_data, header=None, index=None, func_args=None, replace=False, setas=None)¶
Append a column of data or inserts a column to a datafile instance.
- Parameters:
column_data (
numpy.arrayor list or callable) – Data to append or insert or a callable function that will generate new data- Keyword Arguments:
header (string) – The text to set the column header to, if not supplied then defaults to ‘col#’
index (index type) – The index (numeric or string) to insert (or replace) the data
func_args (dict) – If column_data is a callable object, then this argument can be used to supply a dictionary of function arguments to the callable object.
replace (bool) – Replace the data or insert the data (default)
setas (str) – Set the type of column (x,y,z data etc - see
Stoner.Core.DataFile.setas)
- Returns:
self – The
DataFileinstance with the additional column inserted.
Note
Like most
DataFilemethods, this method operates in-place in that it also modifies the original DataFile Instance as well as returning it.
- append(value)¶
S.append(value) – append value to the end of the sequence
- asarray()¶
Provide a consistent way to get at the underlying array data.
- clear() None. Remove all items from D.¶
- closest(value, xcol=None)¶
Return the row in a data file which has an x-column value closest to the given value.
- Parameters:
value (float) – Value to search for.
- Keyword Arguments:
xcol (index or None) – Column in which to look for value, or None to use setas.
- Returns:
ndarray – A single row of data as a
Stoner.Core.DataArray.
Notes
To find which row it is that has been returned, use the
Stoner.Core.DataArray.iindex attribute.
- column(col)¶
Extract one or more columns of data from the datafile.
- Parameters:
col (int, string, list or re) – is the column index as defined for
DataFile.find_col()- Returns:
(ndarray) – One or more columns of data as a
numpy.ndarray.
- columns(not_masked=False, reset=False)¶
Iterate over the columns of data int he datafile.
- count(value=None, axis=0, col=None)¶
Count the number of un-masked elements in the
DataFile.- Keywords:
- valiue (float):
Value to count for
- axis (int):
Which axis to count the unmasked elements along.
- col (index, None):
Restrict to counting in a specific column. If left None, then the current ‘y’ column is used.
- Returns:
(int) – Number of unmasked elements.
- del_column(col=None, duplicates=False)¶
Delete a column from the current
DataFileobject.- Parameters:
col (int, string, iterable of booleans, list or re) – is the column index as defined for
DataFile.find_col()to the column to be deleted- Keyword Arguments:
duplicates (bool) – (default False) look for duplicated columns
- Returns:
self – The
DataFileobject with the column deleted.
Note
If duplicates is True and col is None then all duplicate columns are removed,
if col is not None and duplicates is True then all duplicates of the specified column are removed.
- If duplicates is False and col is either None or False then all masked coplumns are deleeted. If
col is True, then all columns that are not set i the
setasattrobute are deleted.
If col is a list (duplicates should not be None) then the all the matching columns are found.
If col is an iterable of booleans, then all columns whose elements are False are deleted.
- If col is None and duplicates is None, then all columns with at least one elelemtn masked
will be deleted
- del_nan(col=None, clone=False)¶
Remove rows that have nan in them.
- eyword Arguments:
- col (index types or None):
column(s) to look for nan’s in. If None or not given, use setas columns.
- clone (boolean):
if True clone the current object before running and then return the clone not self.
- Returns:
self (DataFile) – Returns a copy of the current object (or clone if *clone*=True)
- del_rows(col=None, val=None, invert=False)¶
Search in the numerica data for the lines that match and deletes the corresponding rows.
- Parameters:
col (list,slice,int,string, re, callable or None) – Column containing values to search for.
val (float or callable) –
- Specifies rows to delete. Maybe:
None - in which case the col argument is used to identify rows to be deleted,
a float in which case rows whose columncol = val are deleted
or a function - in which case rows where the function evaluates to be true are deleted.
a tuple, in which case rows where column col takes value between the minimum and maximum of the tuple are deleted.
- Keyword Arguments:
invert (bool) – Specifies whether to invert the logic of the test to delete a row. If True, keep the rows that would have been deleted otherwise.
- Returns:
self – The current
DataFileobject
Note
If col is None, then all rows with masked data are deleted
if col is callable then it is passed each row as a
DataArrayand if it returns True, then the row will be deleted or kept depending on the value of invert.If val is a callable it should take two arguments - a float and a list. The float is the value of the current row that corresponds to column col abd the second argument is the current row.
Todo
Implement val is a tuple for deletinging in a range of values.
- dir(pattern=None)¶
Return a list of keys in the metadata, filtering with a regular expression if necessary.
- extend(values)¶
S.extend(iterable) – extend sequence by appending elements from the iterable
- filter(func=None, cols=None, reset=True)¶
Set the mask on rows of data by evaluating a function for each row.
- Parameters:
func (callable) – is a callable object that should take a single list as a p[parameter representing one row.
cols (list) – a list of column indices that are used to form the list of values passed to func.
reset (bool) – determines whether the mask is reset before doing the filter (otherwise rows already masked out will be ignored in the filter (so the filter is logically or’d)) The default value of None results in a complete row being passed into func.
- Returns:
self – The current
DataFileobject with the mask set
- find_col(col, force_list=False)¶
Indexes the column headers in order to locate a column of data.shape.
Indexing can be by supplying an integer, a string, a regular expression, a slice or a list of any of the above.
Integer indices are simply checked to ensure that they are in range
String indices are first checked for an exact match against a column header if that fails they are then compiled to a regular expression and the first match to a column header is taken.
A regular expression index is simply matched against the column headers and the first match found is taken. This allows additional regular expression options such as case insensitivity.
A slice index is converted to a list of integers and processed as below
A list index returns the results of feading each item in the list at
find_col()in turn.
- Parameters:
col (int, a string, a re, a slice or a list) – Which column(s) to retuirn indices for.
- Keyword Arguments:
force_list (bool) – Force the output always to be a list. Mainly for internal use only
- Returns:
int, list of ints – The matching column index as an integer or a KeyError
- find_duplicates(xcol=None, delta=1e-08)¶
Find rows with duplicated values of the search column(s).
- Keyword Arguments:
- Returns:
(dictionary of value –
- [list of row indices]):
The unique value and the associated rows that go with it.
Notes
If xcol is not specified, then the
Data.setasattribute is used. If this is also not set, then all columns are considered.
- get(k[, d]) D[k] if k in D, else d. d defaults to None.¶
- get_filename(mode)¶
Force the user to choose a new filename using a system dialog box.
- Parameters:
mode (string) – The mode of file operation to be used when calling the dialog box
- Returns:
str – The new filename
Note
The filename attribute of the current instance is updated by this method as well.
- index(value[, start[, stop]]) integer -- return first index of value.¶
Raises ValueError if the value is not present.
Supporting start and stop arguments is optional, but recommended.
- insert(index, obj)¶
Implement the insert method.
- insert_rows(row, new_data)¶
Insert new_data into the data array at position row. This is a wrapper for numpy.insert.
- Parameters:
row (int) – Data row to insert into
new_data (numpy array) – An array with an equal number of columns as the main data array containing the new row(s) of data to insert
- Returns:
self – A copy of the modified
DataFileobject
- classmethod load(*args, **kargs)¶
Create a new
DataFilefrom a file on disc guessing a better subclass if necessary.- Parameters:
filename (string or None) – path to file to load
- Keyword Arguments:
auto_load (bool) – If True (default) then the load routine tries all the subclasses of
DataFilein turn to load the filefiletype (
DataFile, str) – If not none then tries using filetype as the loader.loaded_class (bool) – If True, the return object is kept as the class that managed to load it, otherwise it is copied into a
Stoner.Dataobject. (Default False)
- Returns:
(Data) – A new instance of
Stoner.Dataor a s subclass ofStoner.DataFileif loaded_class is True.
Note
If filetype is a string, then it is first tried as an exact match to a subclass name, otherwise it is used as a partial match and the first class in priority order is that matches is used.
Some subclasses can be found in the
Stoner.formatspackage.Each subclass is scanned in turn for a class attribute priority which governs the order in which they are tried. Subclasses which can make an early positive determination that a file has the correct format can have higher priority levels. Classes should return a suitable exception if they fail to load the file.
If no class can load a file successfully then a StonerUnrecognisedFormat exception is raised.
- main_data(data_grp)[source]¶
Work through the main data group and build something that looks like a numpy 2D array.
- pop(k[, d]) v, remove specified key and return the corresponding value.¶
If key is not found, d is returned if given, otherwise KeyError is raised.
- popitem() (k, v), remove and return some (key, value) pair¶
as a 2-tuple; but raise KeyError if D is empty.
- remove(value)¶
S.remove(value) – remove first occurrence of value. Raise ValueError if the value is not present.
- remove_duplicates(xcol=None, delta=1e-08, strategy='keep first', ycol=None, yerr=None)¶
Find and remove rows with duplicated values of the search column(s).
- Keyword Arguments:
xcol (index types) – The column)s) to search for duplicates in.
delta (float or array) – The absolute difference(s) to consider equal when comparing floats.
strategy (str, default keep first) –
- What to do with duplicated rows. Options are:
keep first - the first row is kept, others are discarded
average - the duplicate rows are average together.
yerr (ycol,) – When using an average strategey identifies columns that represent values and uncertainties where the proper weighted standard error should be done.
- Returns:
(dictionary of value –
- [list of row indices]):
The unique value and the associated rows that go with it.
Notes
If ycol is not specified, then the
Data.setasattribute is used. If this is also not set, then all columns are considered.
- rename(old_col, new_col)¶
Rename columns without changing the underlying data.
- Parameters:
old_col (string, int, re) – Old column index or name (using standard rules)
new_col (string) – New name of column
- Returns:
self – A copy of the modified
DataFileinstance
- reorder_columns(cols, headers_too=True, setas_too=True)¶
Construct a new data array from the original data by assembling the columns in the order given.
- Parameters:
cols (list of column indices) – (referred to the oriignal data set) from which to assemble the new data set
headers_too (bool) – Reorder the column headers in the same way as the data (defaults to True)
setas_too (bool) – Reorder the column assignments in the same way as the data (defaults to True)
- Returns:
self – A copy of the modified
DataFileobject
- reverse()¶
S.reverse() – reverse IN PLACE
- rolling_window(window=7, wrap=True, exclude_centre=False)¶
Iterate with a rolling window section of the data.
- Keyword Arguments:
- Yields:
ndarray – Yields with a section of data that is window rows long, each iteration moves the marker one row further on.
- rows(not_masked=False, reset=False)¶
Iterate over rows of data.
- save(filename=None, **kargs)¶
Save a string representation of the current DataFile object into the file ‘filename’.
- Parameters:
filename (string, bool or None) – Filename to save data as, if this is None then the current filename for the object is used. If this is not set, then then a file dialog is used. If filename is False then a file dialog is forced.
as_loaded (bool,str) – If True, then the Loaded as key is inspected to see what the original class of the DataFile was and then this class’ save method is used to save the data. If a str then the keyword value is interpreted as the name of a subclass of the the current DataFile.
- Returns:
self – The current
DataFileobject
- search(xcol=None, value=None, columns=None, accuracy=0.0)¶
Search the numerica data part of the file for lines that match and returns the corresponding rows.
- Keyword Arguments:
xcol (index types, None) – a Search Column Index. If None (default), use the current setas.x
value (float, tuple, list or callable, None) – Value to look for
columns (index or array of indices or None (default)) – columns of data to return - none represents all columns.
accuracy (float) – Uncertainty to accept when testing equalities
- Returns:
ndarray – numpy array of matching rows or column values depending on the arguments.
Note
The value is interpreted as follows:
a float looks for an exact match
a list is a list of exact matches
an array or list of booleans (index like Numpy does)
a tuple should contain a (min,max) value.
A callable object should have accept a float and an array representing the value of the search col for the the current row and the entire row.
None opens an interactive span selector in a plot window.
- section(**kargs)¶
Assuming data has x,y or x,y,z coordinates, return data from a section of the parameter space.
- Keyword Arguments:
x (float, tuple, list or callable) – x values ,atch this condition are included inth e section
y (float, tuple, list or callable) – y values ,atch this condition are included inth e section
z (float, tuple,list or callable) – z values ,atch this condition are included inth e section
r (callable) – a
tuple (function that takes a) –
- Returns:
(DataFile) – A
DataFilelike object that includes only those lines from the original that match the section specification
Internally this function is calling
DataFile.search()to pull out matching sections of the data array. To extract a 2D section of the parameter space orthogonal to one axis you just specify a condition on that axis. Specifying conditions on two axes will return a line of points along the third axis. The final keyword parameter allows you to select data points that lie in an arbitrary plane or line. eg:d.section(r=lambda x,y,z:abs(2+3*x-2*y)<0.1 and z==2)
would extract points along the line 2y=3x+2 (note the use of an < operator to avoid floating point rounding errors) where the z-co-ordinate is 2.
- select(*args, **kargs)¶
Produce a copy of the DataFile with only data rows that match a criteria.
- Parameters:
args (various) – A single positional argument if present is interpreted as follows:
If a callable function is given, the entire row is presented to it. If it evaluates True then that row is selected. This allows arbitrary select operations
If a dict is given, then it and the kargs dictionary are merged and used to select the rows
- Keyword Arguments:
kargs (various) –
Arbitrary keyword arguments are interpreted as requestion matches against the corresponding columns. The keyword argument may have an additional __operator* appended to it which is interpreted as follows:
eq value equals argument value (this is the default test for scalar argument)
ne value doe not equal argument value
gt value doe greater than argument value
lt value doe less than argument value
ge value doe greater than or equal to argument value
le value doe less than or equal to argument value
between value lies between the minimum and maximum values of the argument (the default test for 2-length tuple arguments)
ibetween,*ilbetween*,*iubetween* as above but include both,lower or upper values
- Returns:
(DatFile) – a copy the DataFile instance that contains just the matching rows.
Note
if the operator is preceeded by __not__ then the sense of the test is negated.
If any of the tests is True, then the row will be selected, so the effect is a logical OR. To achieve a logical AND, you can chain two selects together:
d.select(temp__le=4.2,vti_temp__lt=4.2).select(field_gt=3.0)
will select rows that have either temp or vti_temp metadata values below 4.2 AND field metadata values greater than 3.
If you need to select on a row value that ends in an operator word, then append __eq in the keyword name to force the equality test. If the metadata keys to select on are not valid python identifiers, then pass them via the first positional dictionary value.
There is a “magic” column name “_i” which is interpreted as the row numbers of the data.
- Example
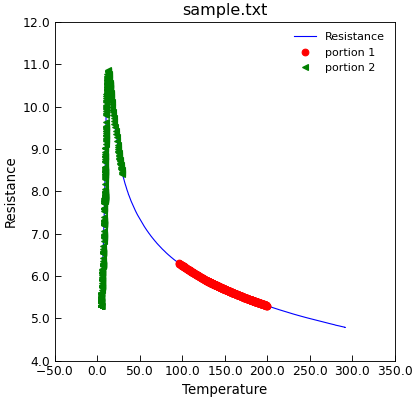
"""Example using select method to pick out data.""" from Stoner import Data d = Data("sample.txt", setas="xy") d.plot(fmt="b-") d.select(Temp__gt=75).select(Res__between=(5.3, 6.3)).plot( fmt="ro", label="portion 1" ) d.select(Temp__lt=30).plot(fmt="g<", label="portion 2")
- setdefault(k[, d]) D.get(k,d), also set D[k]=d if k not in D¶
- sort(*order, **kargs)¶
Sort the data by column name.
- Parameters:
order (column index or list of indices or callable function) – One or more sort order keys.
- Keyword Arguments:
reverse (boolean) – If true, the sorted array isreversed.
- Returns:
(self) – A copy of the
DataFilesorted object
Notes
Sorts in place and returns a copy of the sorted data object fo chaining methods.
If the argument is a callable function then it should take a two tuple arguments and return +1,0,-1 depending on whether the first argument is bigger, equal or smaller. Otherwise if the argument is interpreted as a column index. If a single argument is supplied, then it may be a list of column indices. If no sort orders are supplied then the data is sorted by the
DataFile.setasattribute or if that is not set, then order of the columns in the data.
- split(*args, final='files')¶
Recursively splits the current DataFile into a
Stoner.Folders.DataFolder.- Parameters:
*args (column index or function) – Each argument is used in turn to find key values for the files in the DataFolder
- Keyword Arguments:
final (str) – Controls whether the final argument plaes the files in the DataFolder (default: “files”) or in groups (“groups”)
- Returns:
Stoner.Folders.DataFolder – A
Stoner.Folders.DataFolderobject containing the individualAnalysisMixinobjects
Note
Creates a DataFolder of DataFiles where each one contains the rows from the original object which had the same value of a given column(s) or function.
On each iteration the first argument is called. If it is a column type then rows which amtch each unique value are collated together and made into a separate file. If the argument is a callable, then it is called for each row, passing the row as a single 1D array and the return result is used to group lines together. The return value should be hashable.
Once this is done and the
Stoner.Folders.DataFolderexists, if there are remaining argument, then the method is called recusivelyt for each file and the resulting DataFolder added into the root DataFolder and the file is removed.Thus, when all of the arguments are evaluated, the resulting DataFolder is a multi-level tree.
Warning
There has been a change in the arguments for the split function from version 0.8 of the Stoner Package.
- swap_column(*swp, **kargs)¶
Swap pairs of columns in the data.
Useful for reordering data for idiot programs that expect columns in a fixed order.
- Parameters:
swp (tuple of list of tuples of two elements) – Each element will be iused as a column index (using the normal rules for matching columns). The two elements represent the two columns that are to be swapped.
headers_too (bool) – Indicates the column headers are swapped as well
- Returns:
self – A copy of the modified
DataFileobjects
Note
If swp is a list, then the function is called recursively on each element of the list. Thus in principle the @swp could contain lists of lists of tuples
- to_pandas()¶
Create a pandas DataFrame from a
Stoner.Dataobject.Notes
In addition to transferring the numerical data, the DataFrame’s columns are set to a multi-level index of the
Stoner.Data.column_headersandStoner.Data.setasvalues. A pandas DataFrame extension attribute, metadata is registered and is used to store the metada from the :py:class:1Stoner.Data` object. This pandas extension attribute is in fact a trivial subclass of theStoner.core.typeHintedDict.The inverse operation can be carried out simply by passing a DataFrame into the copnstructor of the
Stoner.Dataobject.- Raises:
**NotImplementedError** if pandas didn't import correctly. –
- unique(col, return_index=False, return_inverse=False)¶
Return the unique values from the specified column - pass through for numpy.unique.
- update([E, ]**F) None. Update D from mapping/iterable E and F.¶
If E present and has a .keys() method, does: for k in E: D[k] = E[k] If E present and lacks .keys() method, does: for (k, v) in E: D[k] = v In either case, this is followed by: for k, v in F.items(): D[k] = v
